Until vaccines and effective therapeutics become available, the practical way to transit safely out of the current lockdown may include the implementation of an effective testing, tracing and tracking system. However, this requires a reliable and clinically validated diagnostic platform for the sensitive and specific identification of SARS-CoV-2.

DNA Barcoding and Diversity Analysis of 19 Economically Important Philippine Sea Cucumbers (Holothuroidea)
This study established the DNA barcodes of 19 economically important Philippine sea cucumbers belonging to Class Holothuroidea under Phylum Echinodermata using the cytochrome c oxidase I (COI) gene.
Early Differential Expression of Galactomannan Biosynthesis Genes in ‘Makapuno’ Coconut (Cocos nucifera L.) revealed by the De Novo Assembly and Analysis of Endosperm Transcriptome
Makapuno is a coconut cultivar with a naturally overproliferating solid endosperm almost filling the nut cavity. At 6-7 months after pollination (MAP), makapuno solid endosperm is phenotypically indistinguishable from the normal type Laguna Tall while at 8-9 MAP makapuno starts to soften and thicken in contrast to the hard and compact appearance of the normal. Here, the expression profiles of 6-7 and 8-9 MAP de novo assembled RNA Sequencing (RNA-Seq) transcriptomes of normal and makapuno were analyzed, and corresponding stages were compared to determine the differentially expressed genes (DEGs).

PGC’s COVID-19 Laboratory is a non-hospital based facility (NHB)
PGC’s Clinical Genomics Laboratory certified to conduct testing for COVID-19 (SARS-CoV-2) by Real-Time PCR is a non-hospital based facility (NHB)
Transcriptome Analysis of ‘Philippine Lono Tall’ Coconut (Cocos nucifera L.) Endosperm Reveals Differential Expression of Genes Involved in Oil Biosynthesis
The ‘Philippine Lono Tall’ (PLNT) is a variant of the more common ‘Philippine Laguna Tall’ (LAGT), which produces fruits with soft endosperm and reported higher fat content. To understand patterns of fatty acid (FA) and oil accumulation in LAGT and PLNT fruits, transcriptomes of 6–7 month-old endosperm samples were analyzed by RNA-Seq. Quantitative PCR was performed to analyze the differential expression of selected genes related to oil biosynthesis. Further, oil samples from the PLNT endosperm were analyzed to determine their FA composition across developmental stages.
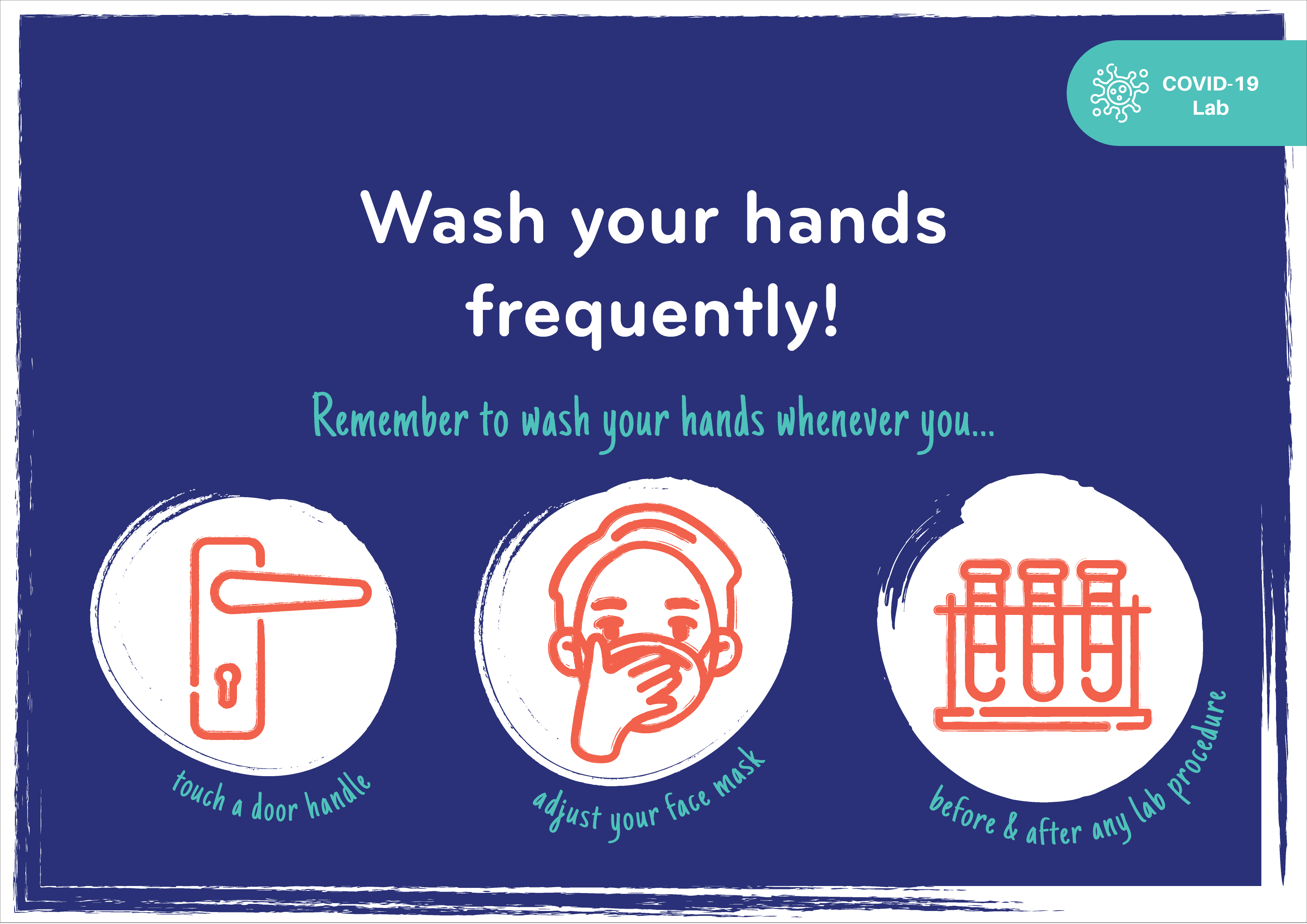
COVID-19 Safety Tips & Precautions
PGC shares its COVID-19 laboratory safety tips and precautions through the illustrations below.

PGC Core Facility for Bioinformatics releases six (6) genome sequences of SARS-CoV-2 from Philippine samples between March 22-28, 2020
The Philippine Genome Center (PGC) thru its Core Facility for Bioinformatics (CFB) releases today to the global community through the GISAID database six (6) genomes of the SARS-CoV-2 sequences from Philippine samples taken in Metro Manila between 26 to 28 March 2020.

PGC’s Clinical Genomics Laboratory certified to conduct testing for COVID-19 (SARS-CoV-2) by Real-Time PCR
Upon completing and passing the Research Institute for Tropical Medicine’s (RITM) proficiency testing for SARS-CoV-2 detection by Real-Time PCR, the Clinical Genomics Laboratory (CGL) of the Philippine Genome Center is now certified to perform independent testing for COVID-19. The certification is released as of April 23, 2020 by the RITM-DOH.

Steps in using the locally made rRT-PCR Detection Kit
How does COVID-19 Testing work? See the general checklist and step-by-step procedure on how the COVID-19 testing works, using the Philippine-made GenAmplify test kit.


