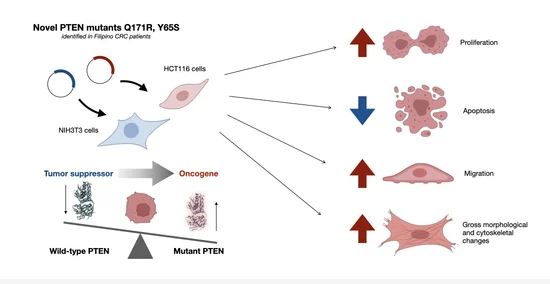
Phosphatase and tensin homolog deleted on chromosome 10, or PTEN, is a well-characterized tumor suppressor with both lipid and protein phosphatase activities. PTEN is often downregulated by epigenetic mechanisms such as hypermethylation, which leads to constitutive activation of the PI3K–Akt pathway.

High rates of tenofovir failure in a CRF01_AE-predominant HIV epidemic in the Philippines
The Philippines has the fastest growing HIV epidemic in the Asia-Pacific. This increase was accompanied by a shift in the predominant HIV subtype from B to CRF01_AE. Increasing evidence points to a difference in treatment responses between subtypes. We examined treatment failure and acquired drug resistance (ADR) in people living with HIV (PLHIVs) after one year on antiretrovirals (ARVs).

PGC Mindanao engages in a collaborative SARS-COV-2 genomics study using a portable sequencing platform
The Oxford Nanopore MinION is a portable sequencer and has the lowest instrument cost among all sequencing platforms with a 97.5 to >99.3% raw read accuracy (Oxford Nanopore Technologies, 2021). The technology has demonstrated its utility for sequencing biological entities, from the simple nano-sized viruses to complex plants and animals, generating sequence information that is sufficient to provide comprehensive insights into the underlying genome architecture. It has also been validated to produce SARS-CoV-2 consensus sequences with the same accuracy as other sequencing platforms (Bull et al., 2020 and Charre et al., 2020).

Incorporating animal forensics in routine meat inspection in the Philippines
Developing mini-barcode primer sets to enhance the utility of conventional techniques is critical in adopting DNA barcoding technology as a robust tool for routine inspections of meat sold
commercially, including those intended for the halal meat industry.
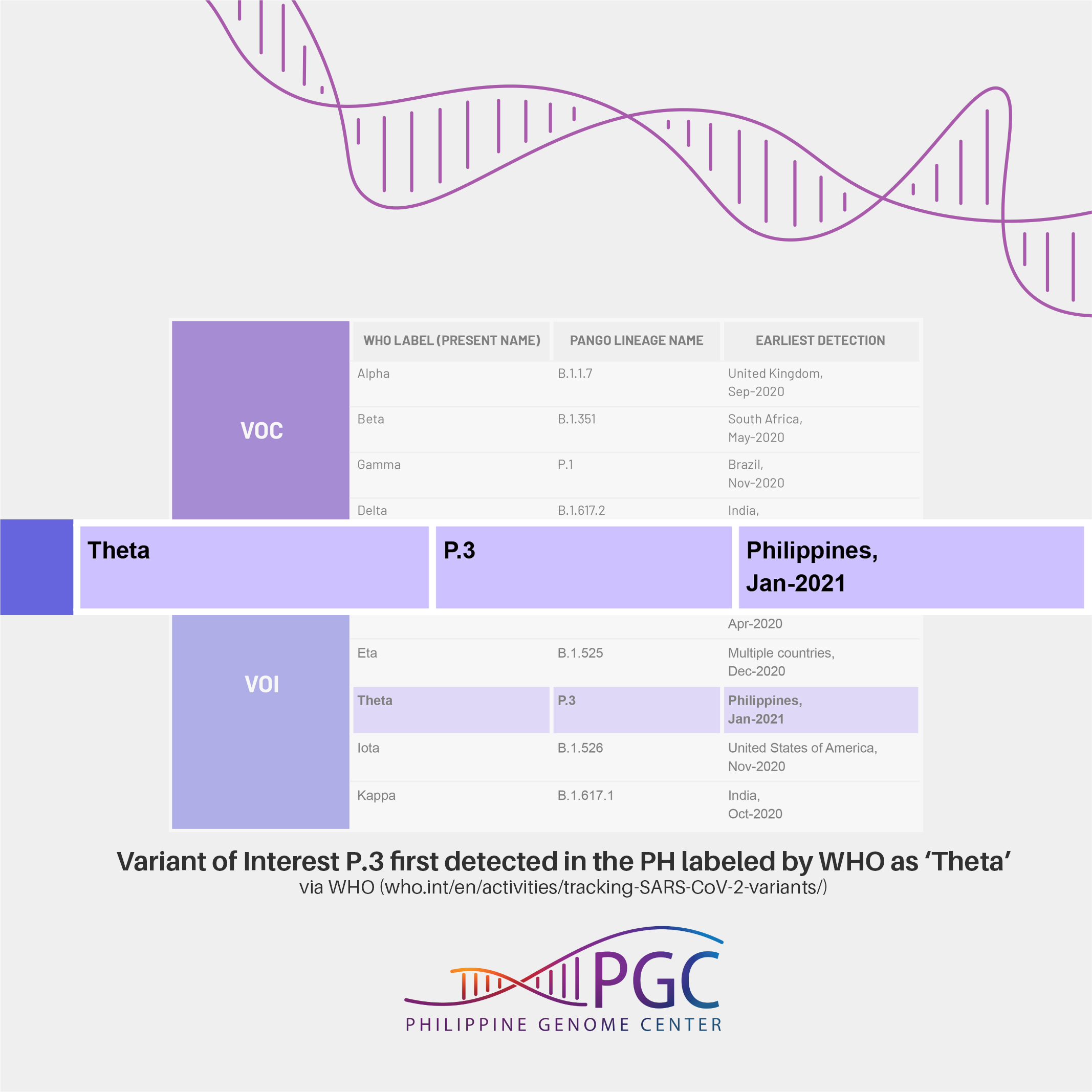
SARS-CoV-2 lineage P.3 first detected in the Philippines labeled as ‘Theta’ by WHO
The recommendation is to use letters of the Greek Alphabet, i.e., Alpha, Beta, Gamma to label the variants. As such, Variant of Interest (VOI) P.3 which was first detected in the Philippines (Jan-21)1 is now labeled by WHO as Theta.
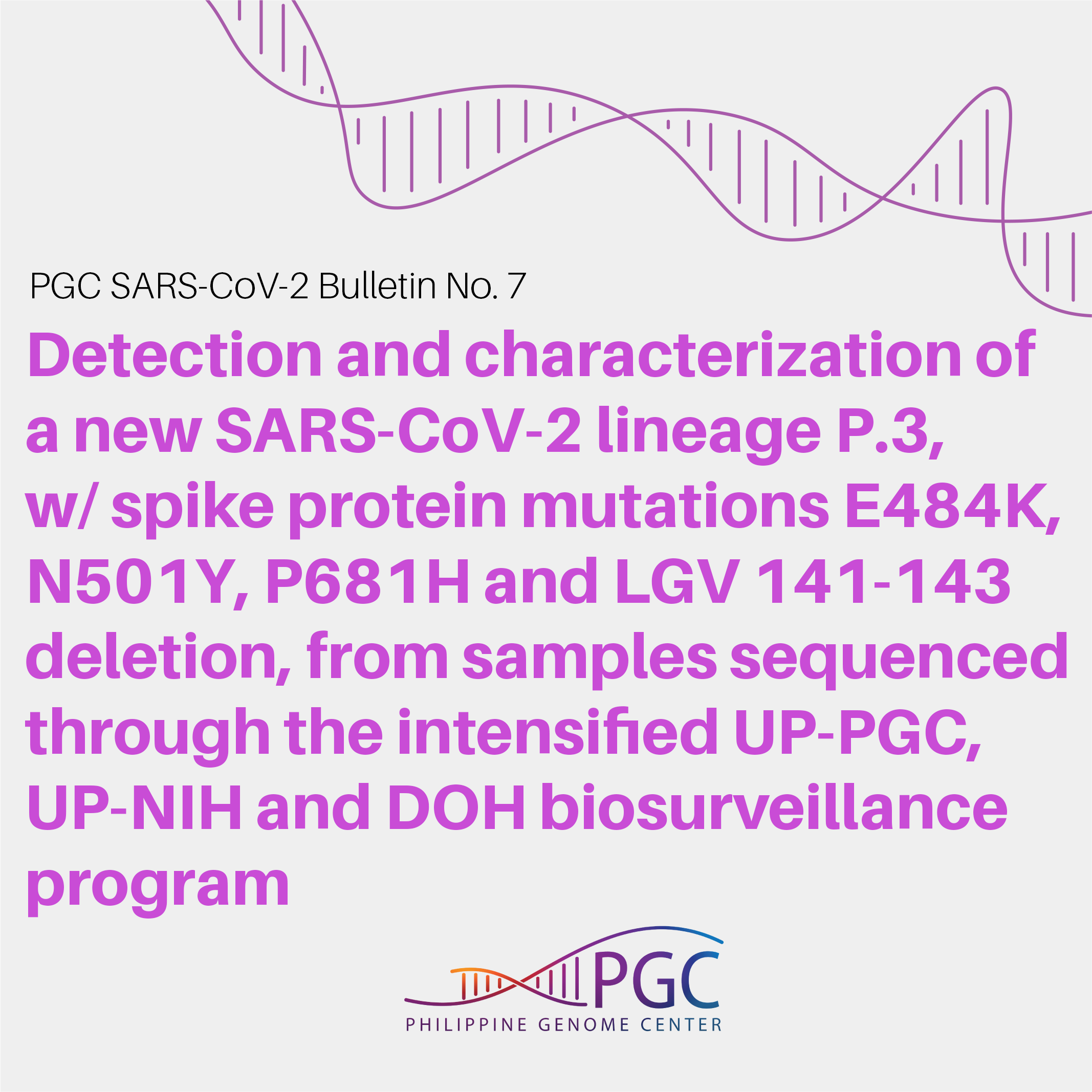
PGC SARS-CoV-2 Bulletin No. 7: Detection and characterization of a new SARS-CoV-2 lineage P.3, with spike protein mutations E484K, N501Y, P681H and LGV 141-143 deletion, from samples sequenced through the intensified UP-PGC, UP-NIH and DOH biosurveillance program
A close examination of the mutation profile of the P.3 viruses revealed that apart from E484K and N501Y, they also share other spike protein mutations that are likely to have functional significance (Figure 2). This includes the P681H mutation, also found in lineage B.1.1.7 viruses, as well as a three-amino acid deletion at positions 141 to 143 (LGV141_143del).
PGC SARS-CoV-2 Bulletin No. 6: First case of the new variant under Lineage B.1.1.7 detected in the Philippines
Through the biosurveillance efforts of the UP – Philippine Genome Center (PGC), in coordination with the Department of Health (DOH) – Epidemiology Bureau and the Inter-Agency Task Force (IATF) Task Force on COVID-19 Variants, we report the first confirmed case of the SARS-CoV-2 B.1.1.7 lineage, more commonly known as the UK variant, in the Philippines. An announcement of this finding can be found in an official DOH press release dated January 13, 2021.

PGC SARS-CoV-2 Bulletin No. 5: New Variants from UK and South Africa NOT detected in 305 local viral samples
Recently, new SARS-CoV-2 variants were detected in the United Kingdom and South Africa with multiple spike protein mutations that may cause substantial changes in certain properties of the virus. These observations spurred biosurveillance efforts in different countries, with some countries outside of the UK and South Africa already reporting the presence of these new variants within their territories. The data presented in this report is part of our own continuing biosurveillance efforts to track the entry of these new variants in the country, as well as other viral mutations that may be of concern locally.

OnePGC: Luzon, Visayas, and Mindanao Virtual Conference 12.10.2020 #PGCParaSaBayan
Philippine Genome Center’s virtual conference on December 10, 2020. More information about this event: https://bit.ly/Program_1PGC2020 OnePGC 2020 phgenome Communications Team