To strengthen the partnerships among the collaborating institutions, a two-week joint-training session was held at the Tokyo Institute of Technology from January 23 to February 3, 2023. The aim of the training was to finalize and optimize a common analysis protocol across the three collaborating teams.

Advisory: Reduced operations on October 19-21, 2022
Please be advised that the respective core facilities (DSCF, CFB, PPMF, CGL) and the administrative services department of the Philippine Genome Center will be implementing reduced operations on October 19-21, 2022 for its annual strategic planning for operational efficiency and team building.
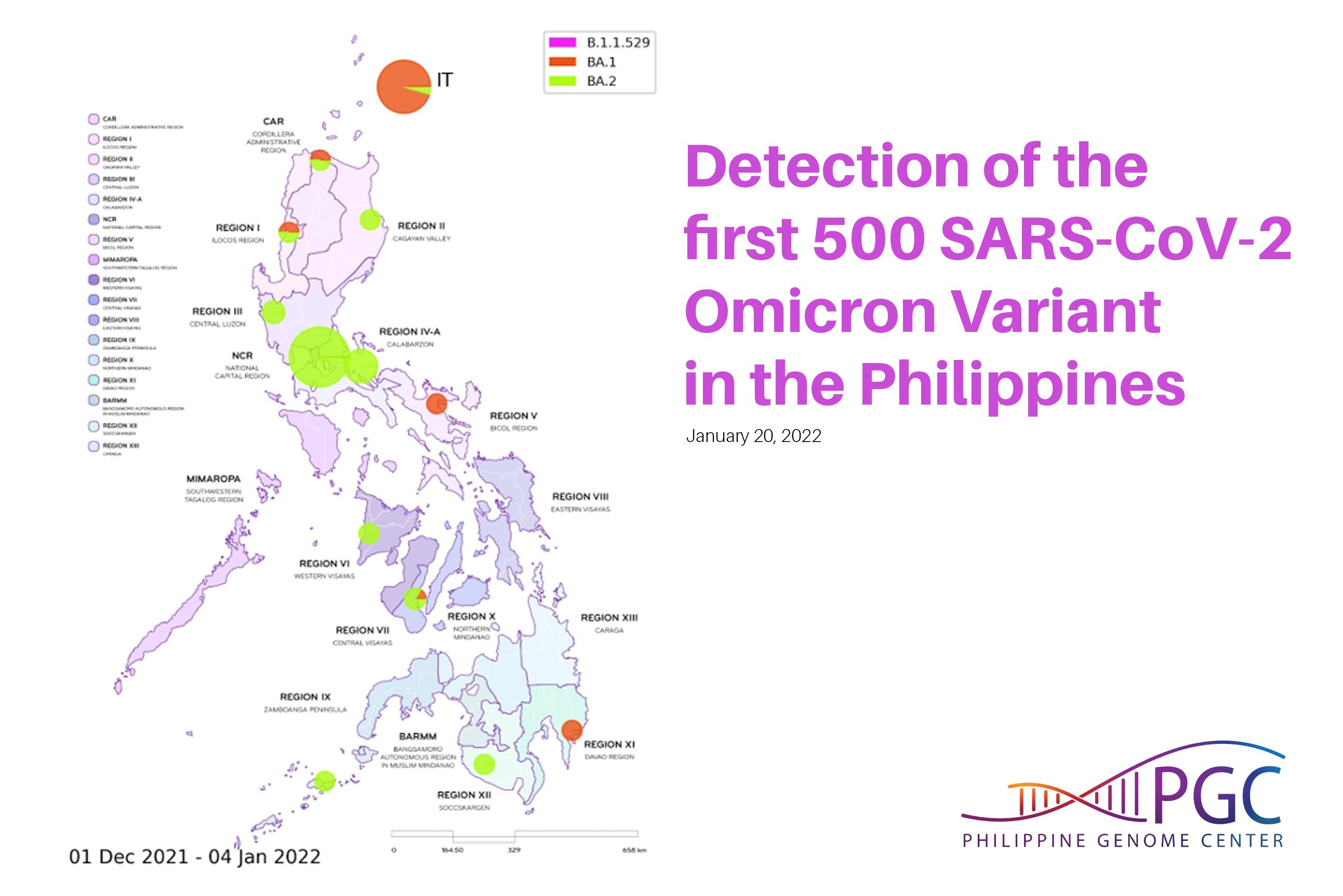
PGC SARS-CoV-2 Bulletin No. 8: Detection of the first 500 SARS-CoV-2 Omicron Variant in the Philippines
This variant, with its more than 30 mutations in the Spike region is hypothesized to be over twice as infectious and twice more likely to escape current vaccines than the Delta variant. Currently, Omicron consists of four (4) phylogenetic lineages: the main lineage B.1.1.529 and its three (3) sublineages: BA.1, BA.2, and BA.3. Based on global statistics, BA.1 is the predominant Omicron lineage being detected worldwide, but a recent increase in the proportion of BA.2 cases has also been observed.

GxB CON 2021: Overcoming Challenges, Building Opportunities
The Philippine Genome Center (PGC) is hosting the Genomics and Bioinformatics Conference 2021 on December 7-9, themed Overcoming Challenges, Building Opportunities—the 3-day conference will consist of talks from experts in health, agriculture, livestock, and fisheries as well as in biodiversity, ethnicity, and forensics.
DOST, UP to inaugurate cutting-edge analytical facility for proteomics research
The new facility under the supervision of Dr. Neil Andrew D. Bascos, has been preparing for this much-awaited launch since last year. Together with his team of young researchers, they have been working on standardizing and optimizing laboratory protocols, and establishing research networks, to promote the use of multi-Omics technologies throughout the Philippines.

Response to the 09 June 2021 statement of Dr. Maximilian Larena and Prof. Mattias Jakobson on the conduct of their Genomic Study on the Filipino Past
Since the Philippine Genome Center is mentioned and we have received numerous queries from colleagues in the country and the region, we are compelled to issue this Statement to address the following:
– the possible inference (from the way the Statement is written) that the head of the Philippine Genome Center’s (PGC) Program on Forensics and Ethnicity connived with the National Commission on Indigenous Peoples (NCIP) and Philippine Health Ethics Research Board (PHREB) against Dr. Larena and Prof. Jakobsson’s research or is responsible for their actions; and
– the imputation that the ethical issues raised by these agencies and PGC aim to discredit “good research” because the head of PGC’s Program on Forensics and Ethnicity is pursuing similar genetic research.

PGC Mindanao engages in a collaborative SARS-COV-2 genomics study using a portable sequencing platform
The Oxford Nanopore MinION is a portable sequencer and has the lowest instrument cost among all sequencing platforms with a 97.5 to >99.3% raw read accuracy (Oxford Nanopore Technologies, 2021). The technology has demonstrated its utility for sequencing biological entities, from the simple nano-sized viruses to complex plants and animals, generating sequence information that is sufficient to provide comprehensive insights into the underlying genome architecture. It has also been validated to produce SARS-CoV-2 consensus sequences with the same accuracy as other sequencing platforms (Bull et al., 2020 and Charre et al., 2020).

PGC Mindanao detects presence of African Swine Fever (ASF) Virus in the region
As the first ever omics facility in Mindanao, PGC Mindanao aims to conduct advance and molecular genomic tests in Mindanao as its contribution to the government response in managing and controlling the Emerging and reemerging livestock diseases.
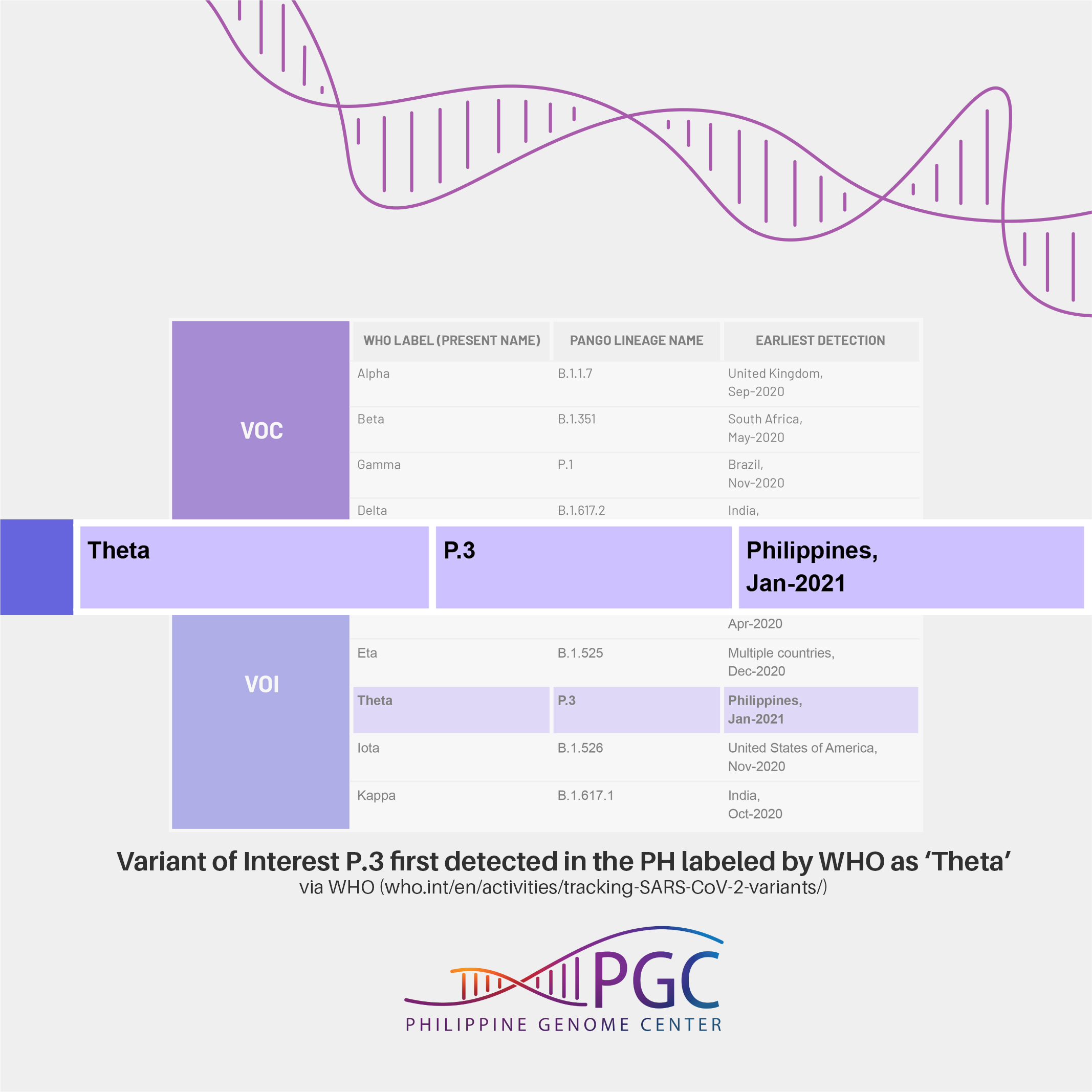
SARS-CoV-2 lineage P.3 first detected in the Philippines labeled as ‘Theta’ by WHO
The recommendation is to use letters of the Greek Alphabet, i.e., Alpha, Beta, Gamma to label the variants. As such, Variant of Interest (VOI) P.3 which was first detected in the Philippines (Jan-21)1 is now labeled by WHO as Theta.
In Vitro Lipid-lowering Properties of the Fruits of Two Bignay [Antidesma bunius (L.) Spreng] Cultivars as Affected by Maturity Stage and Thermal Processing
Bignay [Antidesma bunius (L). Spreng] fruit contains an array of polyphenols and information on how these bioactive compounds vary with cultivar type, maturity stage, and process treatment are unclear. Also, the effects of these variations on the lipid-lowering potential of this Philippine indigenous berry have not been reported.

