The new facility under the supervision of Dr. Neil Andrew D. Bascos, has been preparing for this much-awaited launch since last year. Together with his team of young researchers, they have been working on standardizing and optimizing laboratory protocols, and establishing research networks, to promote the use of multi-Omics technologies throughout the Philippines.

High rates of tenofovir failure in a CRF01_AE-predominant HIV epidemic in the Philippines
The Philippines has the fastest growing HIV epidemic in the Asia-Pacific. This increase was accompanied by a shift in the predominant HIV subtype from B to CRF01_AE. Increasing evidence points to a difference in treatment responses between subtypes. We examined treatment failure and acquired drug resistance (ADR) in people living with HIV (PLHIVs) after one year on antiretrovirals (ARVs).

Response to the 09 June 2021 statement of Dr. Maximilian Larena and Prof. Mattias Jakobson on the conduct of their Genomic Study on the Filipino Past
Since the Philippine Genome Center is mentioned and we have received numerous queries from colleagues in the country and the region, we are compelled to issue this Statement to address the following:
– the possible inference (from the way the Statement is written) that the head of the Philippine Genome Center’s (PGC) Program on Forensics and Ethnicity connived with the National Commission on Indigenous Peoples (NCIP) and Philippine Health Ethics Research Board (PHREB) against Dr. Larena and Prof. Jakobsson’s research or is responsible for their actions; and
– the imputation that the ethical issues raised by these agencies and PGC aim to discredit “good research” because the head of PGC’s Program on Forensics and Ethnicity is pursuing similar genetic research.

PGC Mindanao engages in a collaborative SARS-COV-2 genomics study using a portable sequencing platform
The Oxford Nanopore MinION is a portable sequencer and has the lowest instrument cost among all sequencing platforms with a 97.5 to >99.3% raw read accuracy (Oxford Nanopore Technologies, 2021). The technology has demonstrated its utility for sequencing biological entities, from the simple nano-sized viruses to complex plants and animals, generating sequence information that is sufficient to provide comprehensive insights into the underlying genome architecture. It has also been validated to produce SARS-CoV-2 consensus sequences with the same accuracy as other sequencing platforms (Bull et al., 2020 and Charre et al., 2020).
High Performance Computing and Data Science Workflow in Julia
HPC and Data Science Workflow in Julia #PGCTalks Date: July 16, 2021, Friday, 4:30-6:00pm This webinar is open to the public and registration is free. Abstract Julia is specifically designed from the start of its conception as a language for high-performance computation but at the same time highly interactive. To achieve this, Julia is one […]

Incorporating animal forensics in routine meat inspection in the Philippines
Developing mini-barcode primer sets to enhance the utility of conventional techniques is critical in adopting DNA barcoding technology as a robust tool for routine inspections of meat sold
commercially, including those intended for the halal meat industry.

PGC Mindanao detects presence of African Swine Fever (ASF) Virus in the region
As the first ever omics facility in Mindanao, PGC Mindanao aims to conduct advance and molecular genomic tests in Mindanao as its contribution to the government response in managing and controlling the Emerging and reemerging livestock diseases.
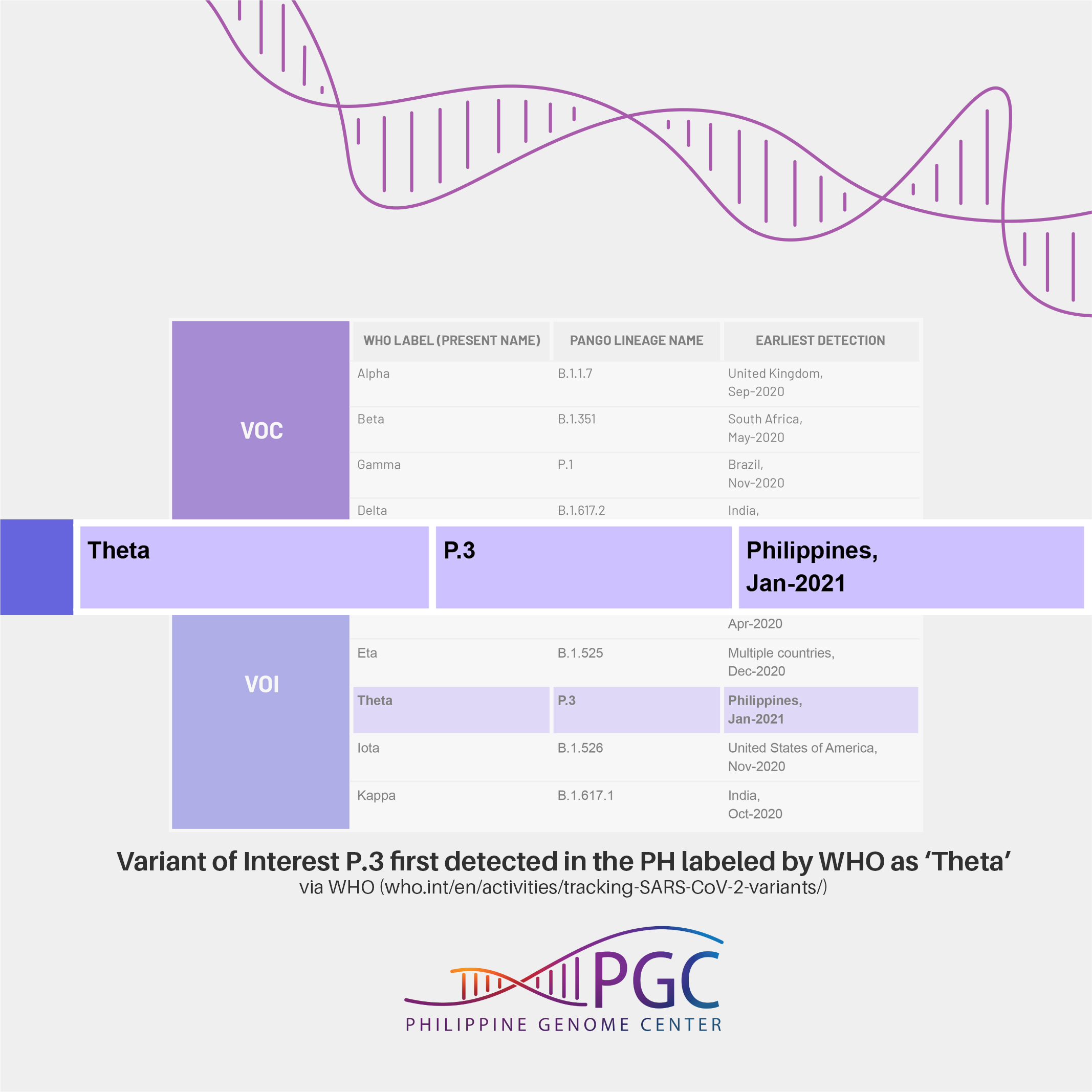
SARS-CoV-2 lineage P.3 first detected in the Philippines labeled as ‘Theta’ by WHO
The recommendation is to use letters of the Greek Alphabet, i.e., Alpha, Beta, Gamma to label the variants. As such, Variant of Interest (VOI) P.3 which was first detected in the Philippines (Jan-21)1 is now labeled by WHO as Theta.
Genetic Structure and Diversity of Banana Bunchy Top Virus (BBTV) in the Philippines
Banana bunchy top virus (BBTV) is an important disease of banana in the Philippines and in other banana-producing countries. This study was conducted to investigate the genetic structure and diversity of Philippine BBTV isolates which remain unexplored in the country. BBTV-infected plant tissues were sampled from banana-growing provinces (i.e., Cagayan, Isabela, Quirino, Batangas, Laguna, Rizal, Quezon, Palawan, Cebu, Leyte, and Davao del Sur) and the partial DNA-R gene of BBTV was sequenced.
Detection and Genome Sequencing of SARS-CoV-2 Variants Belonging to the B.1.1.7 Lineage in the Philippines
Coronavirus disease 2019 (COVID-19) is an infectious disease that has gained pandemic status from the World Health Organization, with millions of cases and deaths recorded worldwide. This global health crisis is caused by the virus referred to as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the genus Betacoronavirus (Coronaviridae), together with the causative agents of the first SARS outbreak in 2003 and the Middle East respiratory syndrome (MERS) in 2012.