A group of researchers from the U.S. Army Medical Directorate–Armed Forces Research Institute of Medical Sciences, the University of the Philippines Manila, and the V. Luna Medical Center recently reported 23 new SARS-CoV-2 genome sequences from the Philippines, all of which were from cases of local transmission (Velasco et al., 2020). Among these samples, one was collected in early April while the rest were obtained in the months of June and July.
Although the Philippine Genome Center (PGC) was not part of the original study, PGC’s Core Facility for Bioinformatics gathered the publicly available sequences and incorporated them in a phylogenomic analysis together with previously reported sequence data from PGC and the Research Institute for Tropical Medicine (RITM).
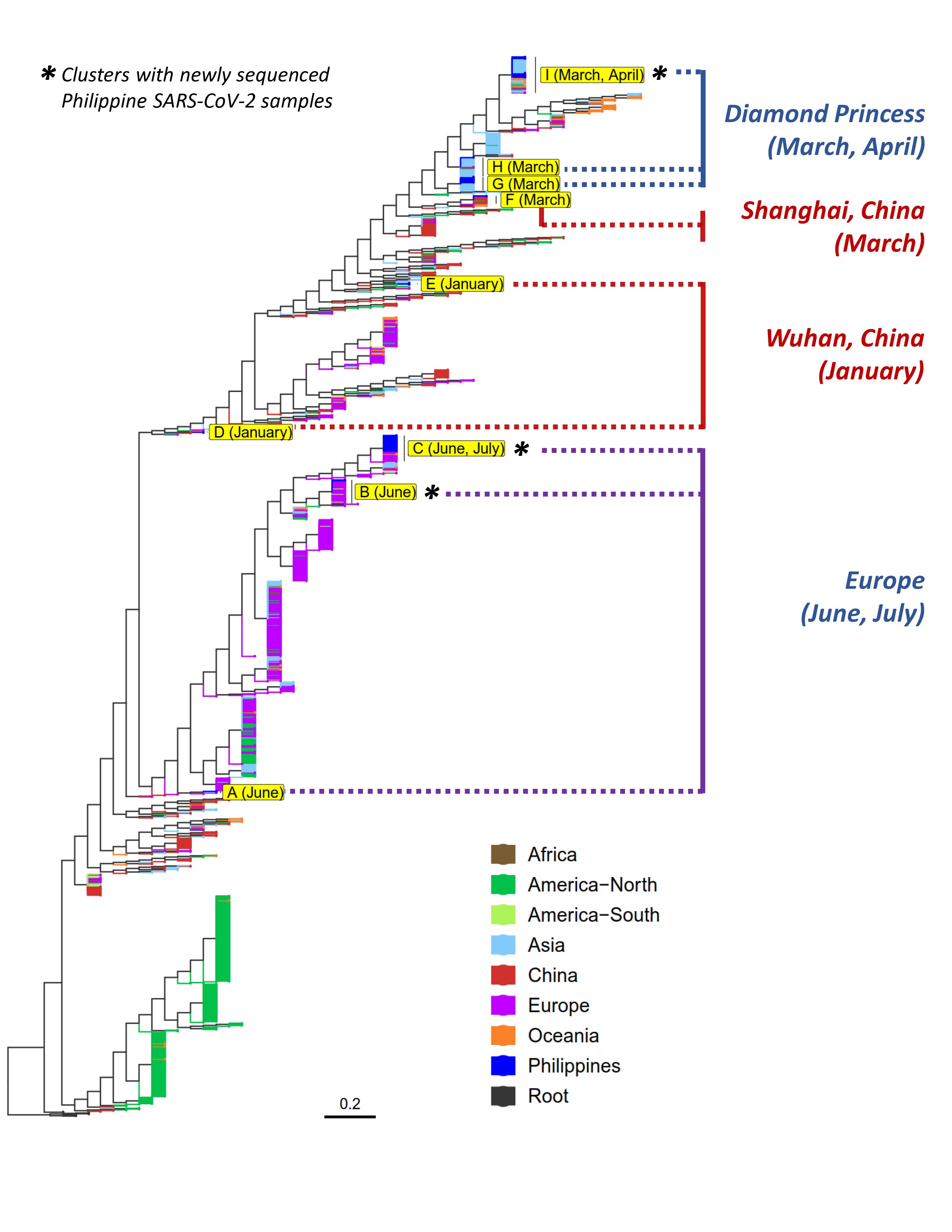
In PGC’s past SARS-CoV-2 bulletins, we reported the detection of the D614G variant in samples from June and July (PGC SARS-CoV-2 Bulletin No. 1) and also inferred the possible sources of infection in the country (PGC SARS-CoV-2 Bulletin No. 2). We earlier hypothesized at least three major waves of viral entry and transmission: (i) January samples linked to foreign visitors from China, (ii) March samples mainly associated with the M/V Diamond Princess Cruise Ship outbreak, and (iii) samples from June onwards that are closely related with European isolates. Here, we report that the analysis results incorporating the 23 newly published Philippine SARS-CoV-2 genome sequences further corroborate our previous observations and hypothesis.
The sample collected in early April clustered mainly with other Philippine isolates collected in March (5 PGC, 4 RITM), as well as with samples from India, Australia, and Japan (Diamond Princess). This particular cluster (Cluster I) is part of a larger group that are mainly linked to the Diamond Princess Cruise Ship cluster of cases. All the other previously reported Philippine samples linked to this outbreak were collected in March. Thus, this is the first evidence pointing to the possibility that the said cluster of cases might have persisted beyond the month of March, amidst the strict quarantine conditions during this period.
In support of our previous findings that SARS-CoV-2 samples from June and July carry the D614G variant and are more closely related to European isolates, the newly sequenced samples collected during the aforementioned months similarly clustered with samples from European countries (Clusters B and C) and were also reported to carry the predominant spike protein variant. Interestingly, majority of the newly sequenced samples grouped closely with one RITM isolate (collected in June) earlier reported to have the D614G mutation as well. These observations suggest that the D614G variant entered the country sometime in June (or possibly late in May) and has since become the predominant SARS-CoV-2 variant in the Philippines, a trend that has also been observed globally.
References
Velasco JM et al. (2020) Coding-complete genome sequences of 23 SARS-CoV-2 samples from the Philippines. Microbiol Resour Announc 9:e01031-20. https://doi.org/10.1128/MRA.01031-20
