The Philippine Genome Center through its Core Facility for Bioinformatics releases today to the global community through the GISAID database six (6) viral genome sequences of the SARS-CoV-2 from COVID-19 cases between March 22-28, 2020 in Metro Manila. All the cases had no travel history outside the Philippines. Two of the sequences reveal that the community-acquired infections cluster closely with samples collected from Japan and Australia from February 15 to March 14, 2020 while four cluster closely with a sample collected from Shanghai, China on February 4, 2020.
The six viral sequences range from 29,869 to 29,981 bases in length similar to the complete SARS-CoV-2 sequences now deposited at the GISAID database, a global database for the sharing of genomic information of all influenza data by researchers around the world. All were sequenced through shotgun viral metagenomics Next Generation Sequencing (NGS) using the High Throughput Illumina sequencers now at the PGC DNA Sequencing Core Facility (DSCF) acquired thru the Commission on Higher Education- Philippine California Advanced Research Institutes (PCARI) Project.
A team of physicians/epidemiologists from the UP National Institutes of Health (NIH) and the Philippine General Hospital (PGH) collected the specimens as part of the DOST-PCHRD sponsored field validation of the GenAmplify rRT-PCR testing kit developed by PGC Deputy Executive Director Raul V. Destura.
The PGC team worked round-the-clock to ensure that the metagenomic assemblies of the viruses are accurate and the sequence variations they obtained are supported by their analysis. The team has also been tracking sequence variations from the local cases of COVID-19 as part of its surveillance of regions targeted by COVID-19 test kits and to see if regions where various vaccines are targeted are mutating or not in local COVID-19 samples. To provide a broader view regarding the sources, spread, and evolution of the SARS-CoV-2 locally, it is important to note that there is still a need to sequence more samples aside from the released six (6) genome sequences.
The viral genome sequences can be accessed at the GISAID database (gisaid.org) with the following accession numbers: PGC001 – EPI_ISL_431833, PGC002 – EPI_ISL_434554, PGC003 – EPI_ISL_434555, PGC004 – EPI_ISL_434556, PGC005 – EPI_ISL_434557, PGC006 – EPI_ISL_434558.
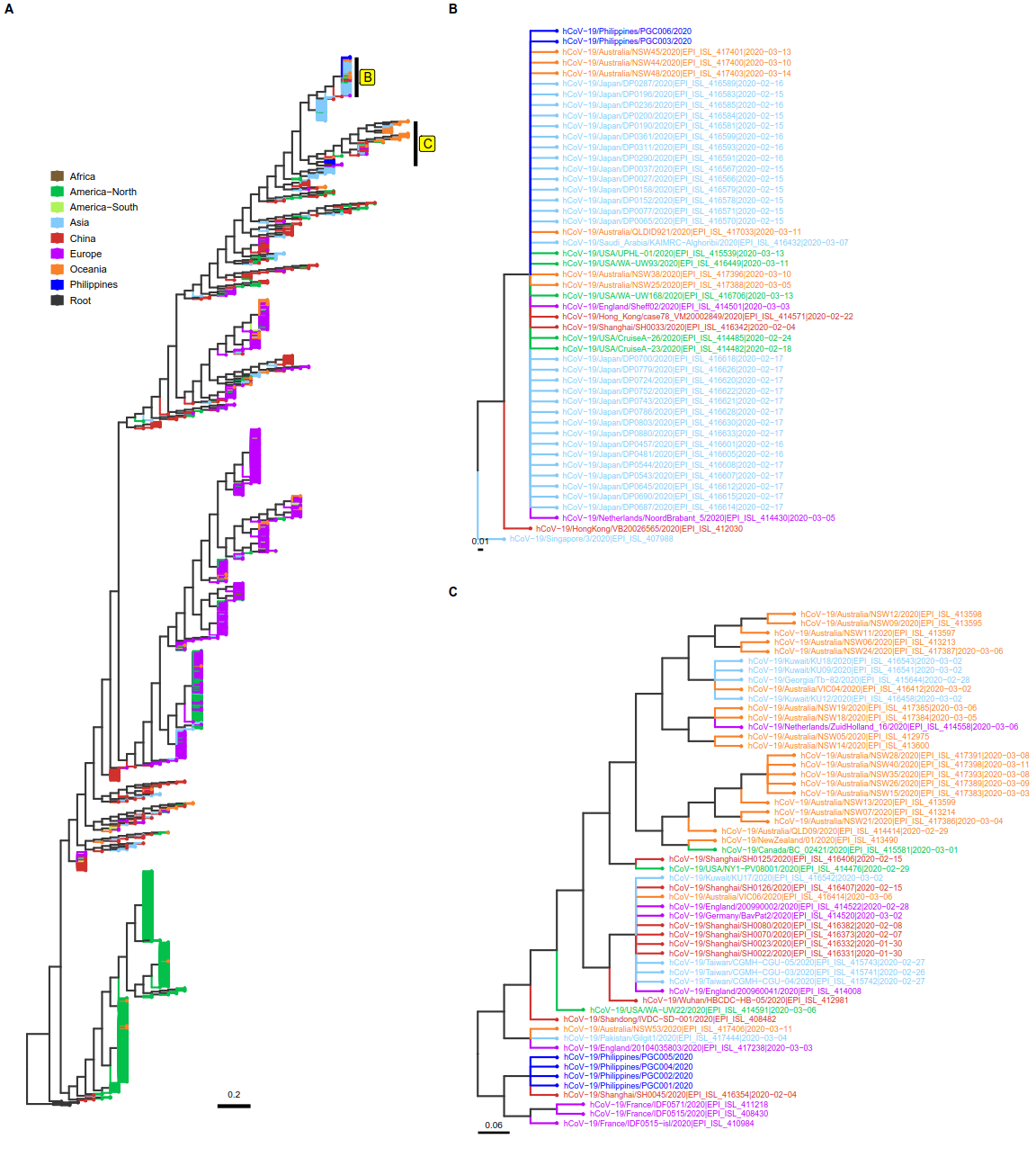 Phylogenetic analysis of SARS-CoV-2 genome sequences. The molecular phylogeny of 1,263 SARS-CoV-2 genome sequences is shown in A. The tips were colored according to geographic locations, mostly corresponding to the continental origins of the isolates. For Asia however, two countries were categorized separately: China which is believed to be the origin of SARS-CoV-2 and the Philippines where the isolates from this study were collected. B and C are subtrees showing the clades where Philippine isolates are clustered.
Phylogenetic analysis of SARS-CoV-2 genome sequences. The molecular phylogeny of 1,263 SARS-CoV-2 genome sequences is shown in A. The tips were colored according to geographic locations, mostly corresponding to the continental origins of the isolates. For Asia however, two countries were categorized separately: China which is believed to be the origin of SARS-CoV-2 and the Philippines where the isolates from this study were collected. B and C are subtrees showing the clades where Philippine isolates are clustered.
