The United Kingdom health ministry has reported a new SARS-CoV-2 variant characterized by multiple mutations in the spike protein region of the virus (Kozlov, 2020). In response to this report, we analyzed Philippine SARS-CoV-2 samples with publicly available genome sequences from the GISAID and NCBI GenBank databases, as well as from internal samples recently collected in Metro Manila as part of the Philippine Genome Center’s biosurveillance efforts. In this bulletin, we present relevant information on the genetic changes found in the UK variant, as well as the results of our latest analysis focusing on locally observed spike protein mutations.
Mutations in the viral spike protein are of particular importance because a large number of these proteins cover the surface of the virus and they facilitate host infection by interacting with another type of surface protein present on human cells known as ACE2 receptors (Huang et al., 2020). Moreover, because spike proteins are the most exposed part of the virus, they are also more likely to trigger the host immune response and are thus important targets for drug and vaccine design (Huang et al., 2020; Walls et al., 2020).
In a report posted at the Global Initiative for Sharing All Influenza Data (GISAID) website, the UK variant is said to harbor multiple spike protein mutations within a single sample, including a combination of the following: H69del, V70del(69), Y145del(143), N501Y, A570D, D614G, P681H(674), T716I, S982A, and D1118H (GISAID, 2020). While the discovery of this new UK variant appears to be concerning, the report cautions that the detailed effects of these mutations remain to be fully determined.
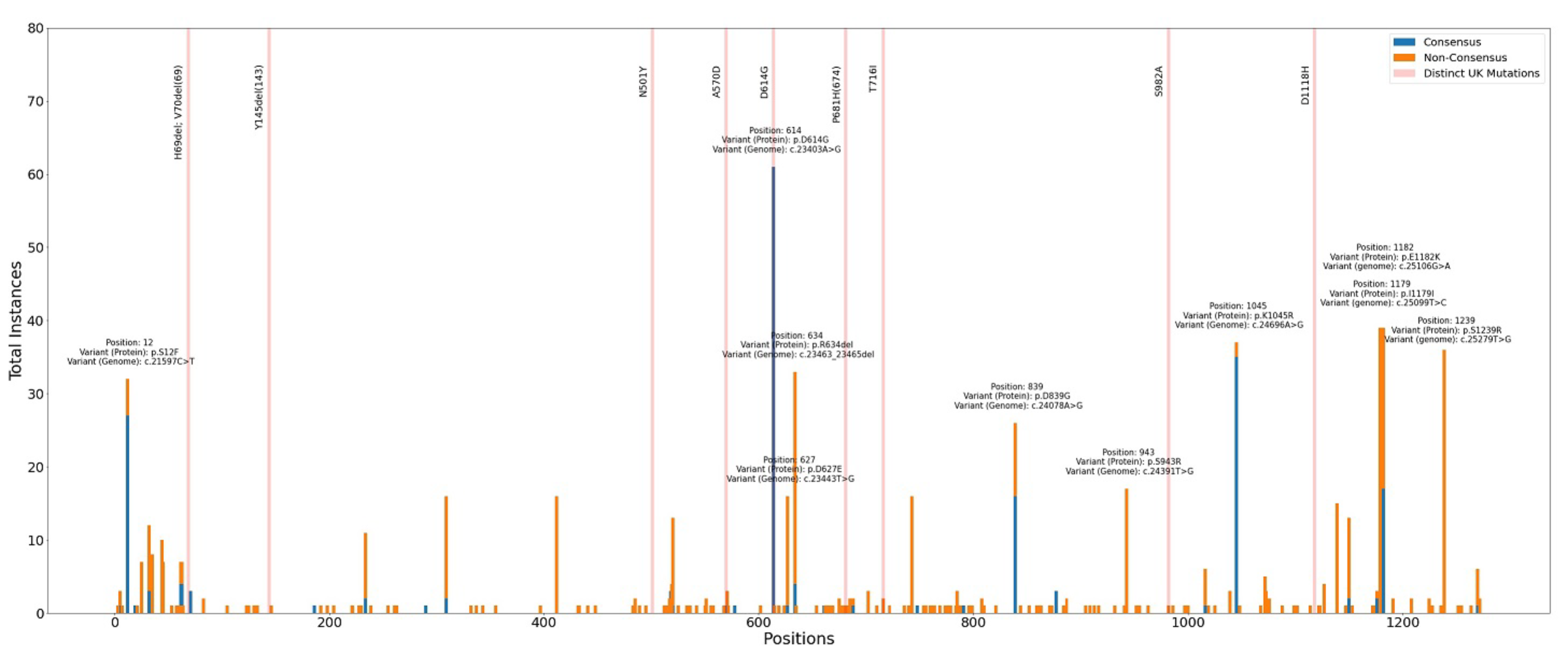
Figure 1. Spike protein mutations found in local SARS-CoV-2 isolates. A total of 65 Philippine SARS-CoV-2 isolate sequences were gathered from publicly available sequence databases (GISAID and NCBI GenBank), as well as from in-house samples recently collected in Metro Manila as part of the Philippine Genome Center’s biosurveillance efforts. The plot shows the number of samples wherein a particular spike protein variant has been observed. Consensus mutations (blue bar) refer to genetic changes found at high (>50%) intrahost frequencies, while non-consensus mutations (orange bar) are those observed at relatively low (≤50%) intrahost frequencies. Typically, mutations with high intrahost frequencies are more likely to be dominant and have higher chances of spreading. The defining mutations of the UK variant are also shown (pale red lines) for comparison. Practically no overlaps were observed between the spike protein mutations of the UK variant and the local SARS-CoV-2 isolates apart from the previously reported D614G mutation. The ten most abundant local spike protein mutations are also labeled, with D614G still appearing to be the most frequently observed.
Based on the results of our latest analysis (Figure 1), we found practically no overlapping spike protein mutations between the described UK variant and the local samples apart from the previously reported D614G mutation (PGC SARS-CoV-2 Bulletin No. 1) – suggesting that the local isolates are not closely related with the UK variant. Although a different set of spike protein mutations was identified locally, majority of the most frequently observed variant positions have already been seen in samples from other countries. Moreover, the current body of information is not enough to determine whether or not these mutations indeed affect viral transmissibility and clinical outcomes. In the meantime, PGC will continue to analyze more SARS-CoV-2 isolates in order to inform and guide our relevant government agencies and the general public regarding significant changes in both locally and globally circulating viral populations.
References
GISAID (2020) UK reports new variant, termed VUI 2020212/01. https://www.gisaid.org/references/gisaid-in-the-news/uk-reports-new-variant-termed-vui-20201201/
Huang Y., et al. (2020) Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologiva Sinica. 41: 1141-1149. https://www.nature.com/articles/s41401-020-0485-4
Kozlov M. 2020. New SARS-CoV-2 variant spreading rapidly in UK. The Scientist. https://www.the-scientist.com/news-opinion/new-sars-cov-2-variant-spreading-rapidly-in-uk-68292
PGC SARS-CoV-2 Bulletin No. 1 (2020) Philippine Genome Center reports detection of the D614G variant of SARS-CoV-2 virus in the Philippines. https://pgc.up.edu.ph/pgc-sars-cov-2-bulletin-no-1-philippine-genome-center-reports-detection-of-the-d614g-variant-of-sars-cov-2-virus-in-the-philippines/
Walls AC., et al. (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 183(16): 1735. https://doi.org/10.1016/j.cell.2020.02.058
