Funded by: PCAARRD
Program Leaders: Cynthia P. Saloma, PHD | Edgar A. Amar, PHD | Leobert D. dela Pena, PHD

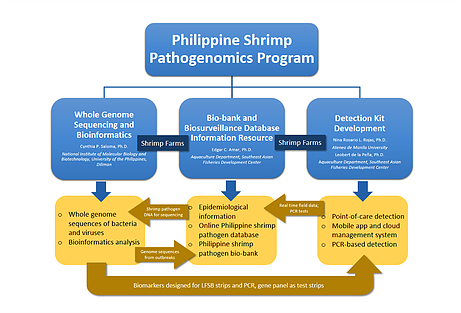
The Shrimp Pathogenomics Program aims to develop a Shrimp Pathogen Bio-bank, an Online Shrimp Pathogen Information Resource, a country-wide biosurveillance system and a pathogenomics initiative to sequence the entire genomes of at least 300 shrimp pathogens with the goal of developing diagnostic tools for the biosurveillance and management of shrimp diseases in the country.
PHILIPPINE PATHOGENOMICS INITIATIVE REVEALS COUNTRYWIDE SITES OF VIBRIOSIS AND AHPND OUTBREAKS IN 2014-2015 AND IDENTIFIES NEW BACTERIAL ISOLATES AFFECTING LOCAL P. monodon and P. vanamei IN AQUACULTURE SHRIMP FARMS
Aquaculture is an important food-producing sector in the Philippines and contributes significantly to the country’s food security, employment, and export earnings. Crustaceans such as shrimp were among the major exports. However, disease outbreaks remain a major constraint and threat to aquaculture production and trade. For the continued viability and competitiveness of the country’s aquaculture sector, strategies need be developed to mitigate and manage infections and respond to outbreaks as well as to track emerging shrimp pathogens nationwide.
In the 1990’s disease outbreaks attributed to luminescent vibriosis occurred in shrimp farms in the Philippines causing Philippine shrimp production to fall. In the country today, while molecular detection tests are available, it is not known whether pathogens isolated from local outbreaks are the same pathogens seen before, or whether they are newly introduced strains or mutated versions of old ones. Genomic information of pathogens is important in improving diagnostic accuracy and in tracing the spread of infection. Genotyping can be applied in aquatic epidemiology to track the source and spread of infection of a trans-boundary pathogen or disease emergence from an environmental reservoir or to determine the pathogen’s local transmission pattern and zoonotic potential.
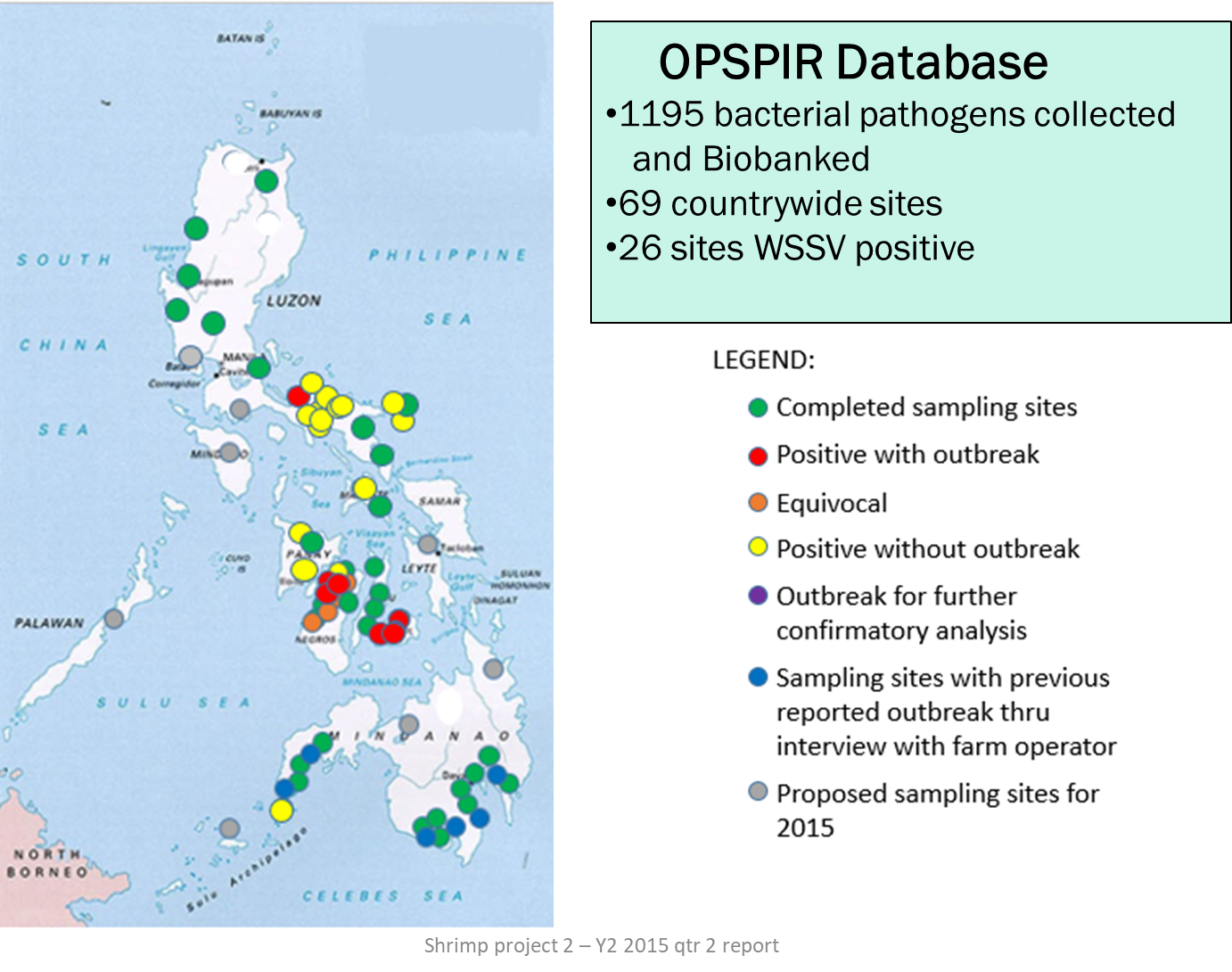
Beginning 2014, we launched the Philippine Shrimp Pathogenomics Program (Fig. 1) to collect, biobank and perform whole genome Next Generation Sequencing (NGS) and develop diagnostic tools of bacterial and viral samples isolated from shrimp tissue from various sites of disease outbreaks in the country (Fig. 2). To date, we have bacterial and viral isolates collected and bio-banked from more than 100 country-wide sites with seven (7) of these sites reported to be AHNPD-positive. Whole genome sequencing of these isolates are done using both Ion PGM and Illumina MiSeq platforms and a bioinformatics analysis workflow has been developed. A website named OPSPIR (Online Philippine Shrimp Pathogen Information Resource) that maps the sites of active as well as of past shrimp disease outbreaks incorporating various epidemiologic information has E. been created. Results of WGS revealed new findings into the bacterial species that predominate vibriosis outbreaks in the Philippines with bacterial populations in these outbreaks found to vary with time and season of the year. We have likewise identified new isotypes of V. parahaemolyticus in diseased shrimp in the country. Further analysis of the whole genome sequences of these shrimp pathogens is expected to result in novel ways to diagnose, prevent, and control shrimp infections. By incorporating and organizing shrimp pathogen genomic information with epidemiologic and other data into a publicly available and accessible online resource, regulatory interventions, diagnostic tests and management practices can be devised for the control and prevention of disease outbreaks affecting the shrimp aquaculture industry in the country.
